| Imputation | Bias (CR) | Coverage (CR) | Bias (SR) | Coverage (SR) |
|---|---|---|---|---|
| normal (-3, -3) | -0.53 | 0.998 | -1.12 | 0.492 |
| normal (0, 0) | -0.12 | 1.000 | -0.71 | 0.886 |
| normal (3, 3) | 0.27 | 1.000 | -0.32 | 0.998 |
| tmvn (1, 5) | -0.04 | 1.000 | -0.01 | 1.000 |
| uniform (0, 0) | -0.16 | 1.000 | -0.75 | 1.000 |
Missing outcomes imputation in bivariate meta-analysis
Padova EMPG Conference, 3 September 2025
Meta-analysis in psychology
A statistical technique to quantitatively summarize the results of multiple studies to derive a more precise effect estimate.
Various sources of heterogeneity: clinical, methodological, statistical
Multivariate meta-analysis
“[…] many clinical studies have more than one outcome variable; this is the norm rather than the exception. These variables are seldom independent and so each must carry some information about the others. If we can use this information, we should.” (Bland 2011)

Multivariate meta-analysis in psychology
Many psychological studies report multiple outcomes (Tyler, Normand, and Horton 2011)
MVMA allows for a joint estimation (Riley et al. 2017)
MVMA handles outcomes missing “by design” (Jackson et al. 2017)
Missing data
Rubin (1976) introduced a foundational framework distinguishing missing data mechanisms: Missing Completely At Random (MCAR), Missing At Random (MAR), Missing Not At Random (MNAR)
These mechanisms differ from missing data patterns, which refer to the observed structure of which values are present or absent.
Patterns = “What” is missing
Mechanisms = “Why” it is missing

Missing Completely At Random
MCAR: Missingness is unrelated to observed or unobserved data. The missing sample is a random subsample.
\[ P(M \mid Y, \phi) = P(M \mid \phi) \]

Missing At Random
MAR: Missingness depends on observed variables.
\[ P(M \mid Y, \phi) = P(M \mid Y_{obs}, \phi) \]

Missing Not At Random
MNAR: Missingness depends on unobserved variables (or both).
\[ P(M \mid Y, \phi) = P(M \mid Y_{mis}, \phi) \]
\[ P(M \mid Y, \phi) = P(M \mid Y_{obs}, Y_{mis}, \phi) \]

Multivariate meta-analysis and sensitivity analysis
What about MNAR outcome measures?
Pre-existing work on imputation of summary-level missing data (Lu 2023; Carpenter, Kenward, and White 2007; Viechtbauer 2022)
Delta-adjustment following multiple imputation of missing outcome (Fiero, Hsu, and Bell 2017)
Core Idea
Replace the unobserved outcomes with many plausible draws, then meta-analyse each imputed dataset and pool the results with Rubin’s rules.
- Assume several donor distributions
- e.g., Uniform, Normal, Multivariate Normal
- Repeated stochastic single imputation
- In each iteration
- Draw missing effect estimates, SEs, and within-study cor
- Fit a bivariate random-effects model (e.g., with
mixmeta) - Store coefficients and vcov
- Combine with Rubin’s rules
- Pool point estimates, between- & within-replicate variance
- Obtain bias, SE, CI coverage across replicates
- Evaluation
- Bias and Coverage
- Sensitivity analysis for the estimates under different assumptions
- Easily interpretable plots with uncertainty intervals
Simulation study
- 50 studies, each with 100 participants
- Two continuous outcomes:
- CR: Clinician Rating
- SR: Self-Report
- Predictors: age, sex, treatment assignment
- Random effects: bivariate normal with \(\rho = 0.7\)
- Estimates obtained via Seemingly Unrelated Regression (SUR) (Zellner 1962; Riley, Tierney, and Stewart 2021)
MNAR mechanism
- Missingness based on effect size estimates:
- CR: more likely missing when treatment effect is low
- SR: more likely missing when treatment effect is high
- Conflict resolution: keep the one furthest from the mean
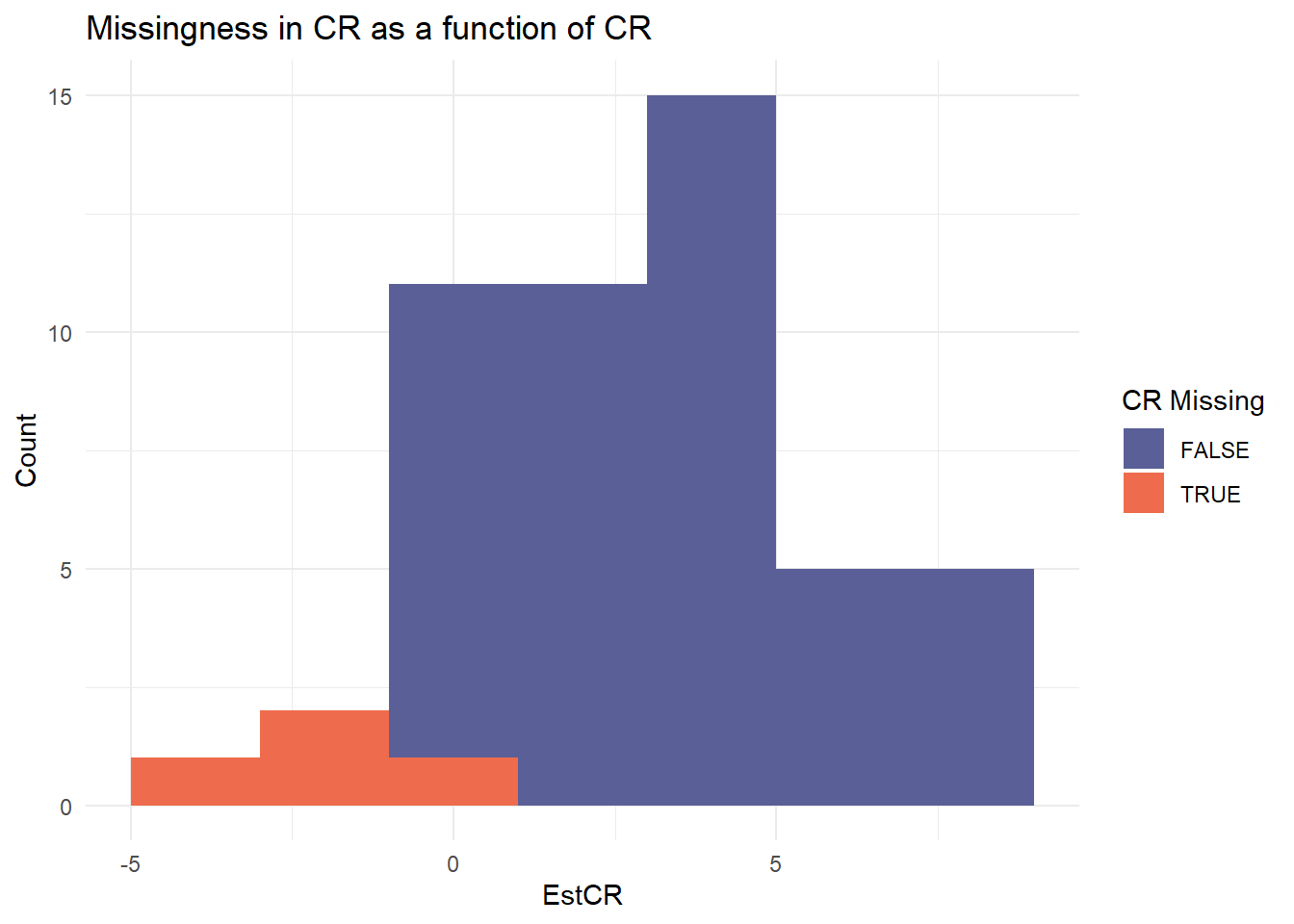 |
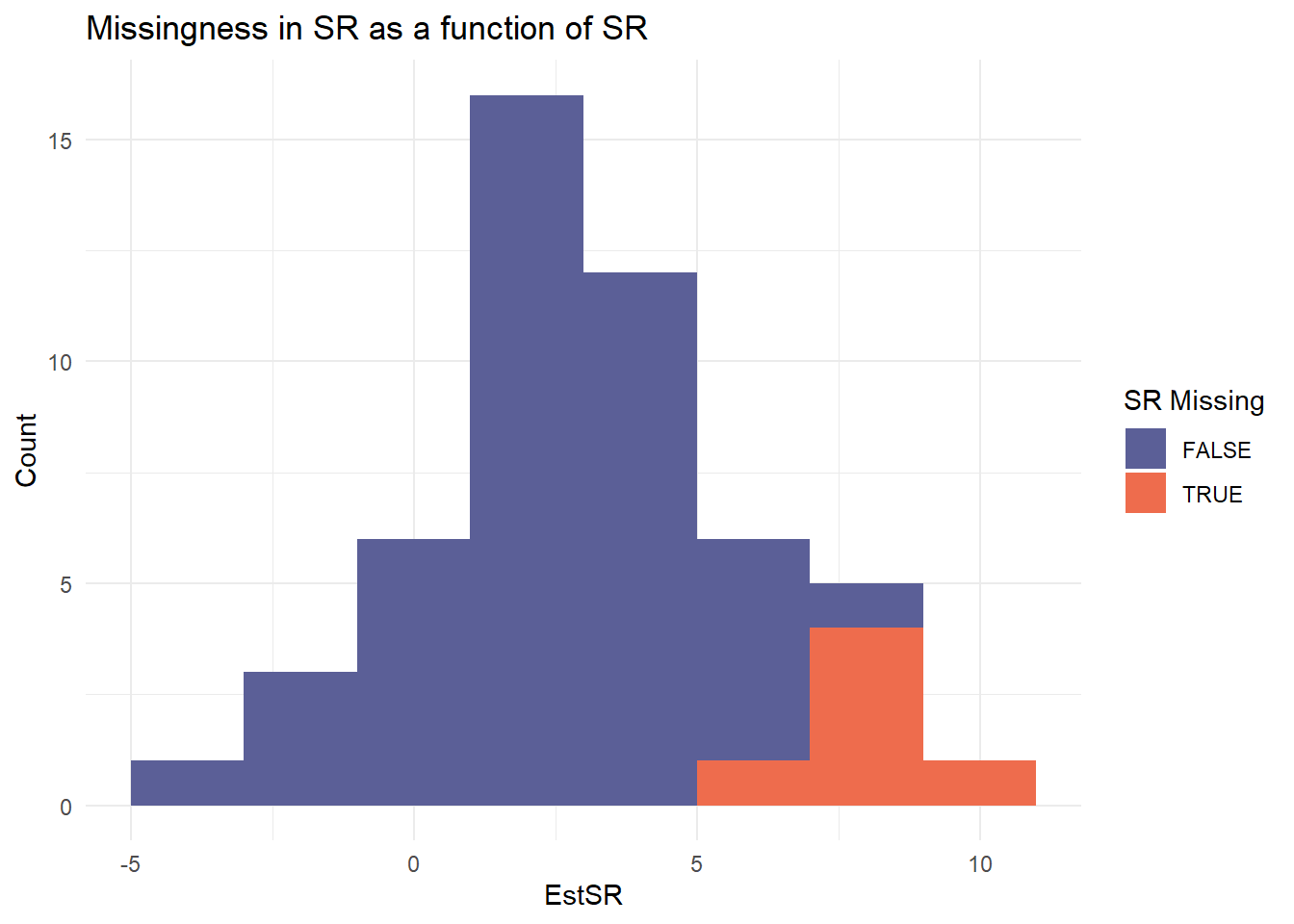 |
Imputation distributions
Missing outcomes were drawn from one of three distributions:
- Normal with mean \(\in {−3, 0, 3}\)
- Truncated multivariate normal (tmvn) with mean = (1, 5)
- Uniform distribution from −100 to 100
Each condition was repeated 10,000 times, with:
- Single stochastic imputation (×100)
- Bivariate random-effects meta-analysis using
mixmeta - Pooled estimates and variances via Rubin’s rule
Estimated CR with 95% CI

Estimated SR with 95% CI

Bias and Coverage for CR and SR
Bias depends on missingness rule
Imputation from uniform distributions are conservative but imprecise
Imputation from truncate multivariate normal best performance, but relies on true model knowledge
In real data multiple assumptions should be tested
Application to real data
Cuijpers et al. (2010) collected data from 48 studies that measure depression on both a clinician rating (HRSD; Hamilton (1960)) and self-report scale (BDI; Beck et al. (1961)). The meta-analysis highlights a substantial difference between the patients’ and clinicians’ evaluations of depression, in favor of the clinician rating.

The dataset
37 studies from Appendix A (HRSD-17 and BDI outcomes, single control group, data from METAPSY database)
3 studies lacked post-treatment data
For multiple comparisons per study:
- Retained the highest mean difference
- BDI used to break ties
# A tibble: 6 × 7
Study N EstCR SECR EstSR SESR Cor.ws
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Arean et al. (1993)a 49 -13.2 1.62 -5.5 1.89 0.7
2 Ayen and Hautzinger (2004)a 21 NA NA -14 1.75 NA
3 Bowers et al. (1993) 16 -3.1 1.52 -5.1 2.83 0.7
4 Bowman, Scogin, and Lyrene (1995)a 20 -7.1 3.17 -10.6 4.86 0.7
5 Brand and Clingempeel (1992) 53 -4.81 1.81 -2.6 2.58 0.7
6 Carpenter et al. (2008) 24 2.10 4.04 NA NA NA Impute missing outcome measures using the package missmeta

Prepare distributions
I make explicit my assumptions regarding missing data, by exploring different scenarios.
library(missmeta)
library(tmvtnorm)
library(mvtnorm)
unif1 <- function(n) runif(n, min = -11, max = 11)
unif2 <- function(n) runif(n, min = -11, max = 11)
norm01 <- function(n) rnorm(n, mean = 0, sd = 6)
norm02 <- function(n) rnorm(n, mean = 0, sd = 6)
norm61 <- function(n) rnorm(n, mean = 6, sd = 6)
norm62 <- function(n) rnorm(n, mean = 6, sd = 6)
normn61 <- function(n) rnorm(n, mean = -6, sd = 6)
normn62 <- function(n) rnorm(n, mean = -6, sd = 6)
sigma <- matrix(c(6^2,
CorCov(6,6,0.7),
CorCov(6,6,0.7),
6^2), nrow = 2)
lower <- c(-100, -100)
upper <- c(100, 100)
mtv1 <- function(n) rtmvnorm(n, mean = c(-6, -6), sigma = sigma, lower = lower, upper = upper)[,1]
mtv2 <- function(n) rtmvnorm(n, mean = c(-6, -6), sigma = sigma, lower = lower, upper = upper)[,2]Impute under different scenarios
Using the function genimp we generate 500 imputed values from each distribution.
imp1 <- list(unif1, norm01, norm61, normn61, mtv1)
imp2 <- list(unif2, norm02, norm62, normn62, mtv2)
out <- mapply(function(i1, i2) {
genimp(
df = dmnar,
iter = 500,
imp1 = i1,
imp2 = i2,
eff1 = "EstCR",
eff2 = "EstSR",
se1 = "SECR",
se2 = "SESR",
cor = "Cor.ws",
N = "N",
imprho = 0.6
)
}, i1 = imp1, i2 = imp2, SIMPLIFY = FALSE)Compute meta-analysis for each imputed dataset
Afterwards, we perform the chosen analyses on each imputed dataset. The user can autonomously choose the preferred meta-analysis package to conduct multivariate meta-analysis (e.g., metaSEM, mixmeta, metafor).
library(mixmeta)
outls = unlist(out, recursive = FALSE)
res = lapply(outls, function(data) {
theta = cbind(data$EstCR, data$EstSR)
Sigma = cbind(data$SECR^2, CorCov(data$SECR, data$SESR, data$Cor.ws), data$SESR^2)
m = mixmeta(theta, S = Sigma, method = "ml")
s = summary(m)
ci = confint(m)
results = data.frame(
eff1 = s$coefficients[1,1],
eff2 = s$coefficients[2,1],
se1 = s$coefficients[1, 2],
se2 = s$coefficients[2, 2],
cov12 = s$vcov[1, 2],
ci.lb1 = ci[1, 1], ci.ub1 = ci[1, 2],
ci.lb2 = ci[2, 1], ci.ub2 = ci[2, 2]
)
})
res <- do.call(rbind, res)
res <- split(res, rep(1:5, each=500))Summarize results for imputed datasets
With makepooldata we prepare the dataset to make it suitable for pooling estimates and standard errors obtained from the iterations using Rubin’s rules.
pooldata <- lapply(res, function(x) {
makepooldata(data = x, effs = "eff", ses = "se", covs = "cov")
})
methods <- c("Uniform (-11, 11)", "Normal CR = 0, SR = 0", "Normal CR = 6, SR = 6",
"Normal CR = -6, SR = -6", "Multivariate Normal (-6, -6)")
sumres <- mapply(function(p, label) {
sumeth_multi(Q_mat = p$Q_mat, U_list = p$U_list, method = label)
}, p = pooldata, label = methods, SIMPLIFY = FALSE)Plot the results

Conclusions
Via a simulation study and a real dataset we have shown the use of a flexible tool for imputing missing outcome data that:
Forces the researcher to make explicit the assumptions on the missing outcome data
Allows for imputation and robustness check of results under different scenarios
Can handle MNAR data
Is easy use and with
missmetaquite forward to implement
Limitations
Results depend on the imputation distributions and parameters
Performance can drop if assumptions are far from reality (SR)
Slow at the moment if one has to test many assumptions
Future work
Increse computational efficiency
Extend to mor than two outcomes in multivariate meta-analysis
Explore proformance against existing imputation methods and sensitivity analysis techniques